1 The Pennsylvania State University, 2 Michigan State University
This article is the second in the series of Fusarium identification tips. Tips for Identifying Fusarium: Part 1: Morphological Identification was published in the May 2021 issue of The Communicator. This article focuses on molecular identification techniques. Morphological identification techniques are especially useful in the diagnostic setting to narrow a given isolate down to a diagnostically meaningful taxon (species, or species complex). However, targeted molecular techniques are generally necessary for accurate species and formae specialis identification. Specifically, obtaining the partial TEF1 DNA sequence described in Geiser et al. (2004) and performing a BLASTn search is emphasized as the method of choice for the broadest range of identification needs. We highlight two important resources currently in pre-print that provide detailed information on sequence-based identification of Fusarium and the updated FUSARIUM-ID v.3.0 database (O’Donnell, et al. 2022; Torres-Cruz, et al. 2022).
The many names of Fusarium Before addressing the details of molecular identification, it is important to understand some of the nomenclatural history of Fusarium so one can better navigate the name changes. When making an identification, searching databases, consulting the literature, and publishing findings, it is crucial to use a naming system that is world-recognized and backed by scientific principles. We recommend following the taxonomic principles outlined in Geiser, et al. (2021) and O’Donnell, et al. (2020) in utilizing the name Fusarium, particularly with regard to the F. solani species complex (FSSC). The Geiser, et al. (2021) proposal is broadly accepted by a global group of plant pathologists and medical mycologists focusing on Fusarium and satisfies both practical and scientific criteria. O’Donnell, et al. (2022) provides guidance on recommended names for taxa.
Can I just sequence the ITS to identify Fusarium?
The availability of DNA sequencing prompted the application of Genealogical Concordance Phylogenetic Species Recognition (GCPSR) to fungi, and it became common practice to define species boundaries through phylogenetic analyses of multiple gene regions (Taylor, et al. 2000). This reinforced the realization that fungal species fitting evolutionary criteria tend to be morphologically cryptic.
Currently, the genus Fusarium contains 23 species complexes, monophyletic lineages consisting of multiple species sharing a common ancestor, all of the descendants of which are members of that group. Within the 23 species complexes of Fusarium there are over 400 “phylospecies” or phylogenetically distinct taxa (O’Donnell, et al. 2022). Some phylospecies have been formally named with Latin binomials, while others have been assigned an ad hoc name that corresponds with multilocus sequence typing (MLST). Phylogenetic studies on species complexes over the years have utilized numerous sequence markers, especially TEF1, RPB1 and RPB2. The ITS and other ribosomal RNA genes such as 28S/LSU rDNA tend to lack resolution at the species level, and in some species complexes may show phylogenetic patterns that do not reflect the organismal history (e.g., FFSC, FOSC: O’Donnell, et al. 1998; O’Donnell, et al. 2009).
The translation elongation factor 1-alpha (TEF1) gene is preferred over ITS for frontline species identification because sequences are usually different enough between closely related taxa to differentiate species, and primers can produce amplicons across the entire range of the genus (Table 1). When the TEF1 sequence is obtained from a Fusarium isolate, it can be used as a query to search using BLAST (basic local alignment tool) against a database of known sequences, such as those contained in the NCBI nucleotide collection (e.g., GenBank). O’Donnell, et al. (2022) uses specific examples to demonstrate some of the nuances of interpreting a TEF1 identification result. When an ITS sequence points toward an unknown being a Fusarium species, it is usually a good idea to follow it up with a TEF1 sequence.
Fusarium-ID v.3.0
Two new articles describing the updated FUSARIUM-ID and current best-practices for sequence-based identification are now available. O’Donnell, et al. (2022) provides details of how to amplify and sequence TEF1 as well as additional loci such as RPB1 and RPB2. The manuscript provides several examples of how to interpret the results of specific BLASTn searches that provide various types of results.
In Torres-Cruz, et al. (2022) the updated Fusarium identification database FUSARIUM-ID v.3.0 is presented with TEF1 sequences from previously unrepresented taxa and all 23 species complexes now included. Metadata for each sequenced isolate have been corrected and standardized across the database. Additionally, the new version is downloadable as a FASTA-formatted text file, so anyone can set up a desktop BLAST database, not relying on the use of the external server which is subject to service interruption. The downloadable database is available at (https://github.com/fusariumid/fusariumid). The manuscript provides detailed tutorials for setting up the database using the commercially available Geneious software package, as well as free options, and provides workflow suggestions for taking sequence results to specific identification.
Other databases for identifying Fusarium
NCBI has a much larger collection of Fusarium sequences, but they may not be connected to available material, and the quality of the taxonomic annotation is mostly up to the depositor. When using a non-curated database to query your sequence, it is important that BLASTn results are carefully considered before settling on an identification. It is ideal to make comparisons against sequences derived from well-characterized isolates, as are available in FUSARIUM-ID. The metadata of the top hits should be consulted to ensure the sequence has not been misidentified, and the current name is employed. O’Donnell, et al. (2015) outlines a workflow and suggestions for interpreting results from GenBank and O’Donnell, et al. (2022) provides detailed examples of navigating such results. One of the advantages of the new, downloadable version of FUSARIUM-ID is that it is easy to add in-house data to the FASTA file, allowing one to search not only against the core database, but all sequences generated at the clinic. It is important, however, for individuals only to share such altered datasets when it is clear to the recipient that it is not the original FUSARIUM-ID, which can only be downloaded from the FUSARIUM-ID github site.
FUSARIOID-ID, maintained by the Westerdijk Fungal Biodiversity Institute in the Netherlands, is another curated database useful for sequence-based identification of Fusarium. It is important to note, however, this database uses different generic names for some Fusarium species, such as those belonging to the Fusarium solani species complex (FSSC) and the Fusarium decemcellulare species complex (FDECSC). Table 2 in O’Donnell, et al. (2022) provides a useful resource for using the favored Fusarium names under the guidance of concepts outlined in Geiser, et al. (2021) and O’Donnell, et al. (2020).
Is my isolate in the genus Fusarium?
Many labs already use ITS primers for identifying fungi. Sequencing ITS and querying it against a curated database or the NCBI nucleotide collection should be sufficient to determine if the unknown isolate belongs to the genus Fusarium or a specific species complex, or even species in some cases. However, if you are able to recognize cultural features consistent with Fusarium it may save you time to go directly to the TEF1 gene and skip ITS.
Which species complex does my isolate belong to?
Diagnosticians must consider the level of specificity that is appropriate for addressing the immediate need of the identification. The host, disease symptoms, the morphological traits of the isolate, and other details will provide the context for determining the precision necessary for making an appropriate diagnosis and management recommendation. The one method that can be consistently applied across the whole genus of Fusarium to determine the species complex of a Fusarium isolate is to query its TEF1 sequence against a taxonomically curated sequence database.
If your BLASTn query of the TEF1 sequence does not return an exact match (or results in “hits” to multiple species complexes) this could mean your isolate cannot be resolved to species or species complex using the TEF1 sequence alone. Each species complex has differing degrees of internal variation and evolutionary relationships with other complexes. Additionally, some species complexes are better represented in some databases than in others. There is not one set of threshold of sequence similarity that can be used to make a sequence-based identification across the genus. This is particularly true for species complexes with recent speciation events, such as F. oxysporum, F. hostae, and F. commune. If results seem ambiguous or queries show results at or below 99.4% (i.e., >4/680 nucleotide differences in TEF1), one may wish to generate sequence data from additional loci and/or compare individual gene trees against a concatenated tree to infer species identity. Phylogenetic analyses of different species complexes have employed different sets of markers, and one may wish to utilize the same markers and data matrices from such studies to provide context for a set of isolates of interest. Otherwise, a combination of TEF1, RPB2, and RPB1, in that order, will usually provide the most taxonomic information.
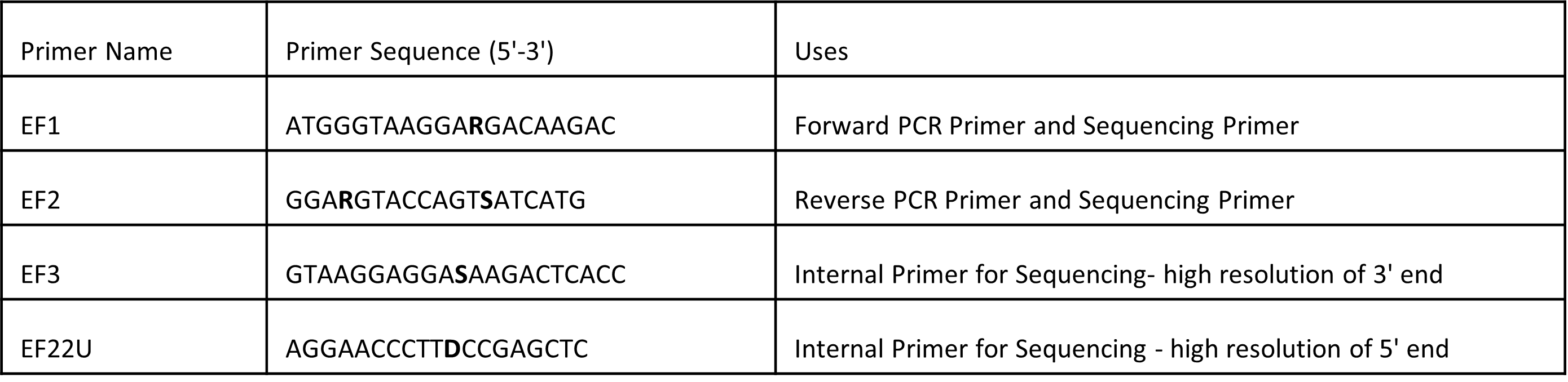
Table 1: Translation Elongation Factor 1-alpha (TEF1) Primers
(adapted from O’Donnell et al. 2022)
Additional tests for specific lineages,
formae speciales, races, etc.
In most cases, a TEF1 sequence alone should be sufficient for determining a species or near-species identification using a BLASTn approach. See O’Donnell, et al. (2022) for guidance regarding ambiguous BLASTn results, which are an inherent phenomenon with this approach. As previously mentioned, it may be necessary to generate sequence data from a second locus and/or perform a GCPSR analysis of closely related species (O’Donnell, et al. 2015). Fusarium is a genus where nonpathogenic strains are commonly found as soil inhabitants or as endophytes or saprophytes in plant tissues. These strains cannot be morphologically distinguished from pathogenic conspecific isolates. Most notably, within the Fusarium oxysporum species complex (FOSC), phylogenetic methods cannot necessarily distinguish nonpathogens from pathogens, even on the same host. Many research groups have developed molecular markers to distinguish specific formae speciales and races (Edel-Hermann and Lecomte 2019). However, formae speciales in F. oxysporum are often the result of multiple evolutionary events, and different races often have separate origins (O’Donnell, et al. 1998). This means targeted diagnostic approaches that cover all race, or distinguish between them, are difficult to develop. The development of molecular assays requires a rigorous validation scheme that evaluates the performance of the test for specificity, sensitivity, and reproducibility. Validation of new diagnostic tests in collaboration with diagnostic laboratories is needed to increase the incorporation of new tests into the routine diagnostic workflow while improving diagnostic accuracy and reliability (Cardwell, et al. 2018).
Some tips on generating sequence data
Sequence data will only be informative and reliable if it is of good quality. The first step to ensuring you generate a quality sequence is to start with a pure culture. Summerell, et al. (2003) and The Fusarium Laboratory Manual (Leslie and Summerell 2006) provide detailed methods and suggested workflows for obtaining a pure Fusarium culture from different substrates. Specialized media is recommended for certain substrates. A previous article in this series, Tips for the purification of fungi and oomycetes, outlines several approaches to obtaining a single-spore culture.
Fortunately, extracting DNA from Fusarium tissue is fairly straightforward and many extraction protocols have been shown to be effective for generating PCR products for Sanger sequencing (Geiser, et al. 2004; O’Donnell, et al. 1997; O’Donnell, et al. 2021). It is important that your initial steps of breaking down the fungal cell walls to release DNA are successful, you can read more on tissue homogenization in a previous P&V tips article. Minimizing PCR inhibitors such as RNA, proteins, etc. is another important consideration that was also covered in a previous article, Tips for coping with PCR inhibitors. A good CTAB DNA extraction miniprep procedure, or a column-based commercial extraction mini kit will generally provide consistent results for downstream PCR and sequencing.
The primers EF1 and EF2 used to amplify the TEF1 regions through PCR can also be used as sequencing primers. When used as sequencing primers, EF1 and EF2 will capture the entirety of the sequence, which is important if you need to generate a phylogeny. One may choose to use separate internal primers for sequencing (O’Donnell, et al. 2022). Most of the identification signal is at the 5’ end of the EF1/EF2 amplicon because it is intron-rich, but the entirely exonic 3’ end provides useful information as well, particularly at the species complex level (O’Donnell, et al. 2022).
The quality of the sequence must be considered, and the raw chromatogram data should be assessed. Fluorescent peaks that are not distinct, have poor signal, or overlap with others should be trimmed and not included in the queried sequence. Re-calling ambiguous sites as ‘N’, and then ignoring those sites in comparison to BLASTn hits, is a good way to avoid mistakenly identifying a sequence as different.
Workshops and Training
The International Fusarium Laboratory Workshop is held annually to provide training in Fusarium identification. This workshop will be held at Kansas State University in June 2022. Visit the workshop website for updates on future workshops.
Resources
Cardwell, K., Dennis, G., Flannery, A.R., Fletcher, J., Luster, D., Nakhla, M., Rice, A., Shiel, P., Stack, J., Walsh, C., and Levy, L. 2018. Principles of diagnostic assay validation for plant pathogens: A basic review of concepts. Plant Health Progress 19:272-278. https://doi.org/10.1094/PHP-06-18-0036-RV
Edel-Hermann, V. and Lecomte, C. 2019. Current status of Fusarium oxysporum formae speciales and races. Phytopathology 109(4): 512-530. https://doi.org/10.1094/PHYTO-08-18-0320-RVW
Geiser, D. M., Jimenez-Gasco, M. M., Kang, S., Makalowska, I., Veeraraghavan, N., Ward, T. J., Zhang, N., Kuldau, G. A., and O’Donnell, K. 2004. FUSARIUM-ID v. 1.0: A DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 110:473–479. https://doi.org/10.1023/B:EJPP.0000032386.75915.a0
Geiser, D. M., Al-Hatmi, A. M. S., Aoki, T., Arie, T., Balmas, V., Barnes, I., Bergstrom, G. C., Bhattacharyya, M. K., Blomquist, C. L., Bowden, R. L., Brankovics, B., Brown, D. W., Burgess, L. W., Bushley, K., Busman, M., Cano-Lira, J. F., Carrillo, J. D., Chang, H.-X., Chen, C.-Y., Chen, W., Chilvers, M. I., Chulze, S., Coleman, J. J., Cuomo, C. A., de Beer, Z. W., de Hoog, G. S., Del Castillo-Múnera, J., Del Ponte, E. M., Diéguez-Uribeondo, J., Di Pietro, A., Edel-Hermann, V., Elmer, W. H., Epstein, L., Eskalen, A., Esposto, M. C., Everts, K. L., Fernández-Pavía, S. P., da Silva, G. F., Foroud, N. A., Fourie, G., Frandsen, R. J. N., Freeman, S., Freitag, M., Frenkel, O., Fuller, K. K., Gagkaeva, T., Gardiner, D. M., Glenn, A. E., Gold, S. E., Gordon, T. R., Gregory, N. F., Gryzenhout, M., Guarro, J., Gugino, B. K., Gutiérrez, S., Hammond-Kosack, K. E., Harris, L. J., Homa, M., Hong, C.-F., Hornok, L., Huang, J.-W., Ilkit, M., Jacobs, A., Jacobs, K., Jiang, C., Jimenez-Gasco, M. M., Kang, S., Kasson, M. T., Kazan, K., Kennell, J. C., Kim, H.-S., Kistler, H. C., Kuldau, G. A., Kulik, T., Kurzai, O., Laraba, I., Laurence, M. H., Lee, T., Lee, Y.-W., Lee, Y.- H., Leslie, J. F., Liew, E. C. Y., Lofton, L. W., Logrieco, A. F., López-Berges, M. S., Luque, A. G., Lysøe, E., Ma, L.-J., Marra, R. E., Martin, F. N., May, S. R., McCormick, S. P., McGee, C., Meis, J. F., Migheli, Q., Mohamed Nor, N. M. I., Monod, M., Moretti, A., Mostert, D., Mulé, G., Munaut, F., Munkvold, G. P., Nicholson, P., Nucci, M., O'Donnell, K., Pasquali, M., Pfenning, L. H., Prigitano, A., Proctor, R. H., Ranque, S., Rehner, S. A., Rep, M., Rodríguez-Alvarado, G., Rose, L. J., Roth, M. G., Ruiz-Roldán, C., Saleh, A. A., Salleh, B., Sang, H., Scandiani, M. M., Scauflaire, J., Schmale, D. G. III, Short, D. P. G., Šišić, A., Smith, J. A., Smyth, C. W., Son, H., Spahr, E., Stajich, J. E., Steenkamp, E., Steinberg, C., Subramaniam, R., Suga, H., Summerell, B. A., Susca, A., Swett, C. L., Toomajian, C., Torres-Cruz, T. J., Tortorano, A. M., Urban, M., Vaillancourt, L. J., Vallad, G. E., van der Lee, T. A. J., Vanderpool, D., van Diepeningen, A. D., Vaughan, M. M., Venter, E., Vermeulen, M., Verweij, P. E., Viljoen, A., Waalwijk, C., Wallace, E. C., Walther, G., Wang, J., Ward, T. J., Wickes, B., Wiederhold, N. P., Wingfield, M. J., Wood, A. K. M., Xu, J.-R., Yang, X.-B., Yli-Matilla, T., Yun, S.-H., Zakaria, L., Zhang, H., Zhang, N., Zhang, S. X., and Zhang, X. 2021. Phylogenomic analysis of a 55.1 kb 19-gene dataset resolves a monophyletic Fusarium that includes the Fusarium solani Species Complex. Phytopathology 111:1064–1079. https://doi.org/10.1094/PHYTO-08-20-0330-LE
Leslie, J.F. and Summerell, B.A. 2006. The Fusarium Laboratory Manual. Blackwell Publishing, Ames, IA.
O’Donnell, K., Cigelnik, E., Weber, N.S., Trappe, J.M. 1997. Phylogenetic relationships among ascomycetous truffles and the true and false morels inferred from 18S and 28S ribosomal DNA sequence analysis. Mycologia 89(1): 48-65. https://doi.org/10.2307/3761172
O'Donnell, K., Cigelnik, E., and Nirenberg, H. I. 1998. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia 90:465–493. https://doi.org/10.2307/3761407
O’Donnell, K., Sutton, D. A., Rinaldi, M. G., Gueidan, C., Crous, P. W., and Geiser, D. M. 2009. Novel multilocus sequence typing scheme reveals high genetic diversity of human pathogenic members of the Fusarium incarnatum-F. equiseti and F. chlamydosporum species complexes within the United States. J. Clin. Microbiol. 47:3851–3861.
https://doi.org/10.1128/JCM.01616-09
O’Donnell, K., Ward, T. J., Robert, V. A. R. G., Crous, P. W., Geiser, D. M., and Kang, S. 2015. DNA sequence-based identification of Fusarium: Current status and future directions. Phytoparasitica 43:583–595. https://doi.org/10.1007/s12600-015-0484-z
O’Donnell, K., Al-Hatmi, A. M. S., Aoki, T., Brankovics, B., Cano-Lira, J. F., Coleman, J. J., de Hoog, G. S., Di Pietro, A., Frandsen, R. J. N., Geiser, D. M., Gibas, C. F. C., Guarro, J., Kim, H.-S., Kistler, H. C., Laraba, I., Leslie, J. F., López-Berges, M. S., Lysøe, E., Meis, J. F., Monod, M., Proctor, R. H., Rep, M., Ruiz-Roldán, C., Šišic ́, A., Stajich, J. E., Steenkamp, E. T., Summerell, B. A., van der Lee, T. A. J., van Diepeningen, A. D., Verweij, P. E., Waalwijk, C., Ward, T. J., Wickes, B. L., Wiederhold, N. P., Wingfield, M. J., Zhang, N., and Zhang, S. X. 2020. No to Neocosmospora: phylogenomic and practical reasons for continued inclusion of the Fusarium solani species complex in the genus Fusarium. mSphere 5:e00810-20. https://doi.org/10.1128/mSphere.00810-20
O’Donnell, K., Laraba, I., and Geiser, D. M. 2021. Pure culture and DNA sequence-based identification of Fusarium from symptomatic plants and diverse substrates. Pages 1-20 in: Fusarium Wilt: Methods and Protocols. J. Coleman, ed. Humana Press, New York.
O’Donnell, K., Whitaker, B. K., Laraba, I., Proctor, R. H., Brown, D. W., Broders, K., Kim, H. -S., McCormick, S. P., Busman, M., Aoki, T., Torres-Cruz, T. J., and Geiser, D. M. 2022. DNA sequence-based identification of Fusarium: A work in progress. (In review). https://doi.org/10.1094/PDIS-09-21-2035-SR
Summerell, B.A., Salleh, B., and Leslie, J. F. 2003. A utilitarian approach to Fusarium identification. Plant Disease 87(2): 117-128. https://doi.org/10.1094/PDIS.2003.87.2.117
Taylor, J. W., Jacobson, D. J., Kroken, S., Kasuga, T., Geiser, D. M., Hibbett, D. S., and Fisher, M. C. 2000. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 31:21–32. https://doi.org/10.1006/fgbi.2000.1228
Torres-Cruz, T. J., Whitaker, B. K., Proctor, R. H., Broders, K., Laraba, I., Kim, H. -S., Brown, D. W., O’Donnell, K., Estrada-Rodriguez, T.L., Lee, Y. -H., Cheong, K., Wallace, E. C., McGee, C. T., Kang, S., Geiser, and D. M. 2022. FUSARIUM-ID v.3.0: An updated, downloadable resource for Fusarium species identification. (In review). https://doi.org/10.1094/PDIS-09-21-2105-SR

